About me
I am an Assistant Professor of Statistics at the University of Chicago. My research develops statistical methods for high-dimensional and multimodal data integration, with applications in biology, environmental science, and public health. I work on structured estimation, graph-constrained models, and graph neural networks, with recent projects including uncertainty quantification for sparse canonical correlation analysis, denoising on graphs, and statistical foundations for GNNs.
📢 NEWS: The group is recruiting!
We welcome applications on a rolling basis for postdocs, rotation PhD students (already accepted to UChicago), and a limited number of MS or advanced undergraduate students. Learn more about our research and available opportunities.
We welcome applications on a rolling basis for postdocs, rotation PhD students (already accepted to UChicago), and a limited number of MS or advanced undergraduate students. Learn more about our research and available opportunities.
- Postdoctoral fellows: Email cdonnat [at] uchicago.edu with your interests and CV.
- UChicago PhD/Masters/Undergrad students: Email your interests, CV, and transcript.
- Note: For those outside UChicago, we may not have the bandwidth to respond.
💼 NEWS: Consulting Availability
I am available for research consulting in areas including statistical methodology, high-dimensional data analysis, graph-based modeling, and data integration for the life sciences. For inquiries, please contact me at cdonnat [at] uchicago.edu.
I am available for research consulting in areas including statistical methodology, high-dimensional data analysis, graph-based modeling, and data integration for the life sciences. For inquiries, please contact me at cdonnat [at] uchicago.edu.
Research Interests
My research develops statistical methodology for the analysis, integration, and uncertainty quantification of high-dimensional and structured data, with a particular emphasis on graphs, networks, and multimodal biological datasets. Graphs provide a unifying framework for representing complex relationships—whether between microbial taxa, cell populations in tissues, or interacting variables in a multivariate system. They differ from traditional Euclidean data in two key ways:
(1) Dependence structure: Nodes are connected, breaking the independence assumptions underlying many classical methods. Inference must explicitly incorporate graph topology, edge weights, and neighborhood structure. (2) Irregularity: Graphs lack a canonical ordering, reference frame, or homogeneity in node roles (degree, centrality, etc.), requiring statistical and machine learning methods that adapt to structural heterogeneity.
Methodological Directions
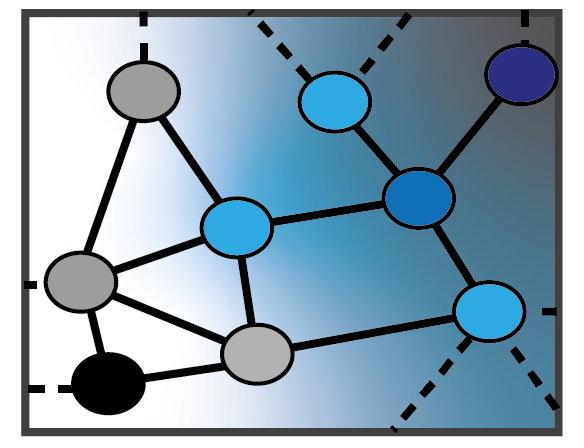
1 — Theory and Methods for Graph Neural Networks
Developing a principled statistical foundation for GNNs, including bias–variance tradeoffs, topology-aware generalization, interpretable architectures, and reliable model selection. Learn more.
Developing a principled statistical foundation for GNNs, including bias–variance tradeoffs, topology-aware generalization, interpretable architectures, and reliable model selection. Learn more.
2 — Structured Estimation and Graph-Constrained Models
Designing algorithms for dimension reduction and matrix factorization that incorporate known network structure or sparsity, with provable guarantees and interpretable outputs.
Designing algorithms for dimension reduction and matrix factorization that incorporate known network structure or sparsity, with provable guarantees and interpretable outputs.
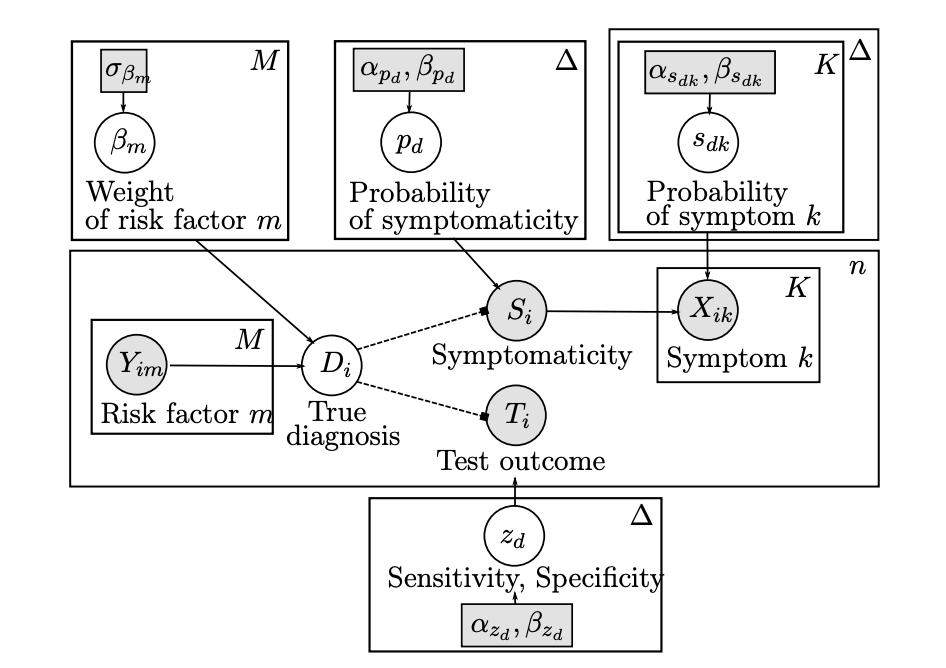
3 — Multimodal Data Integration and Uncertainty Quantification
Integrating heterogeneous datasets (e.g., genomic, transcriptomic, spatial) using sparse CCA, regularized regression, and probabilistic graphical models, with an emphasis on uncertainty assessment.
Integrating heterogeneous datasets (e.g., genomic, transcriptomic, spatial) using sparse CCA, regularized regression, and probabilistic graphical models, with an emphasis on uncertainty assessment.
Applications
- Thermotolerance in photosynthetic microbes — Linking genomic, transcriptomic, and metabolomic data of cyanobacteria and Chlamydomonas to phenotypes under temperature or light stress.
- Family networks and child welfare — Modeling kinship structures to study their impact on outcomes in child protective services.
- Spatial transcriptomics — Detecting spatial gene expression patterns and cell–cell interactions.
- Microbial communities — Modeling environmental and host-associated microbiomes using graph-based approaches.
