Constrained Bayesian ICA for Brain Connectomics
Published in arXiv (Under submission), 2019
- Joint work with Leonardo Tozzi and Susan Holmes*
Brain connectomics is a developing field in neurosciences which strives to understand cognitive processes and psychiatric diseases through the analysis of interactions between brain regions. However, in the high-dimensional, low-sample, and noisy regimes that typically characterize fMRI data, the recovery of such interactions remains an ongoing challenge: how can we discover patterns of co- activity between brain regions that could then be associated to cognitive processes or psychiatric disorders?
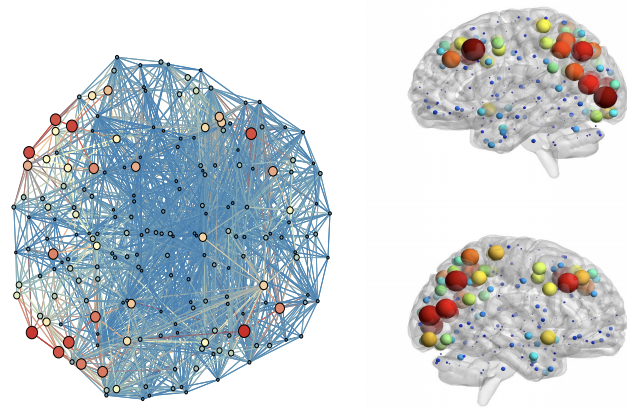
In this paper, we investigate a constrained Bayesian ICA approach which, in comparison to current methods, simultaneously allows (a) the flexible integration of multiple sources of information (fMRI, DTI, anatomical, etc.), (b) an automatic and parameter-free selection of the appropriate sparsity level and number of connected submodules and (c) the provision of estimates on the uncertainty of the recovered interactions. Our experiments, both on synthetic and real-life data, validate the exibility of our method and highlight the the benefits of integrating anatomical information for connectome inference.
Recommended citation: Donnat Claire, Leonardo Tozzi and Susan Holmes*. “Constrained Bayesian ICA for Brain Connectomics” arXiv (2019) — Under submission.
