A Bayesian Hierarchical Network for Combining Heterogeneous Data Sources in Medical Diagnoses
Published in Proceedings of the Machine Learning for Health NeurIPS Workshop, 2020
Joint work with Freddy Bunbury, Nina Miolane and Jack Kreindler.
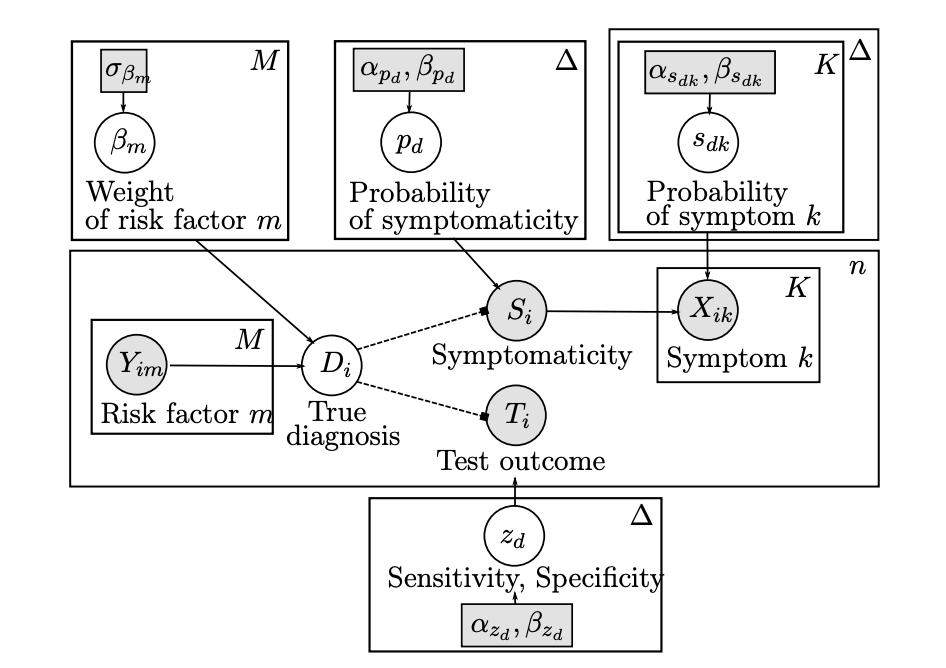
The increasingly widespread use of affordable, yet often less reliable medical data and diagnostic tools poses a new challenge for the field of Computer Aided Diagnosis: how can we combine multiple sources of information with varying levels of precision and uncertainty to provide an informative diagnosis estimate with confidence bounds? Motivated by a concrete application in lateral flow antibody testing, we devise a Stochastic Expectation-Maximization algorithm that allows the principled integration of heterogeneous and potentially unreliable data types. Our Bayesian formalism is essential in (a) flexibly combining these heterogeneous data sources and their corresponding levels of uncertainty, (b) quantifying the degree of confidence associated with a given diagnostic, and (c) dealing with the missing values that typically plague medical data. We quantify the potential of this approach on simulated data, and showcase its practicality by deploying it on a real COVID19 immunity study.
Recommended citation: Donnat, Claire, Nina Miolane, Freddy Bunbury, and Jack Kreindler. “A bayesian hierarchical network for combining heterogeneous data sources in medical diagnoses.” In Machine Learning for Health (pp. 53-84). PMLR..
