Modeling the Heterogeneity in COVID-19’s Reproductive Number and its Impact on Predictive Scenarios
Published in Journal of Applied Statistics, 2020
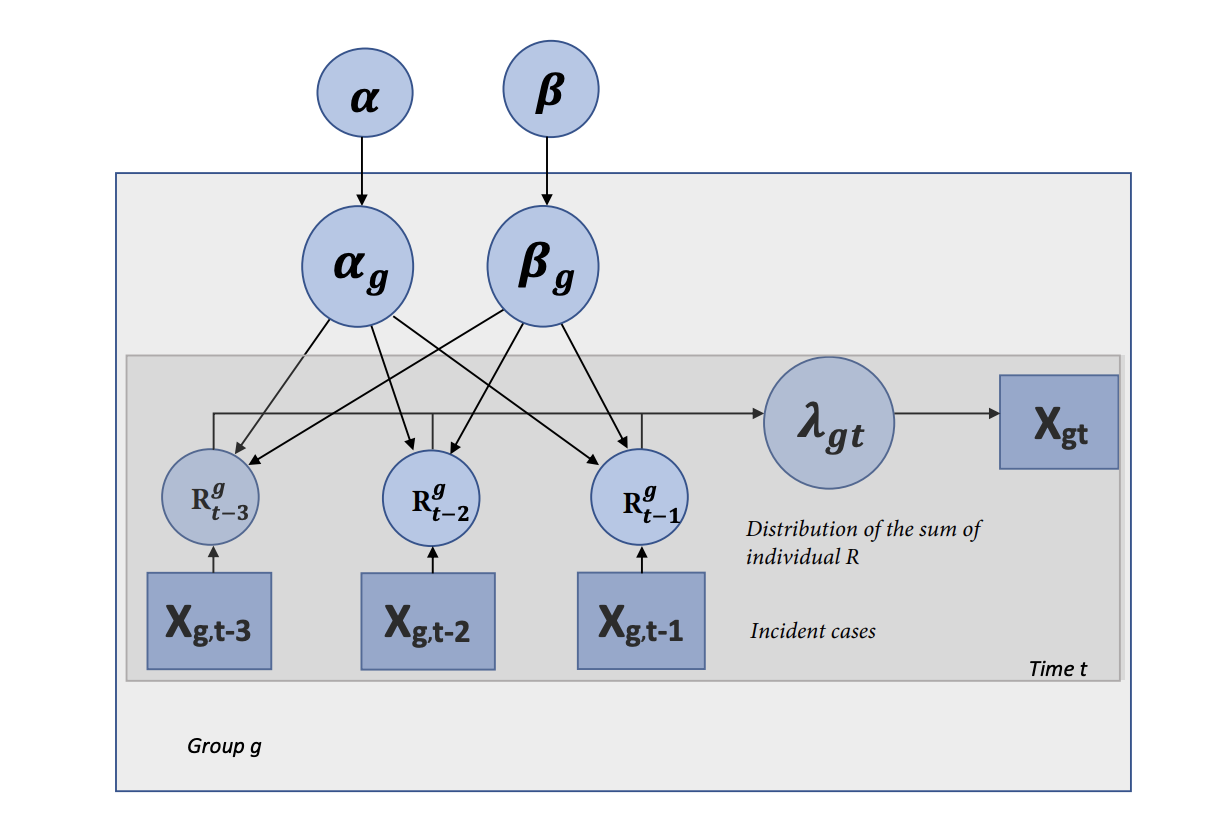
The correct evaluation of the reproductive number R for COVID-19 —which characterizes the average number of secondary cases generated by each typical primary case— is central in the quantification of the potential scope of the pandemic and the selection of an appropriate course of action. In most models, R is modeled as a universal constant for the virus across outbreak clusters and individuals — effectively averaging out the inherent variability of the transmission process due to varying individual contact rates, population densities, demographics, or temporal factors amongst many. Yet, due to the exponential nature of epidemic growth, the error due to this simplification can be rapidly amplified and lead to inaccurate predictions and/or risk evaluation. In this paper, we propose to study this question through a Bayesian perspective, creating a bridge between the agent-based and compartmental approaches commonly used in the literature. After deriving a Bayesian model that captures at scale the heterogeneity of a population and environmental conditions, we simulate the spread of the epidemic as well as the impact of different social distancing strategies, and highlight the strong impact of this added variability on the reported results.
Recommended citation: Donnat Claire, and Susan Holmes. “Modeling the heterogeneity in COVID-19’s reproductive number and its impact on predictive scenarios.” Journal of Applied Statistics (2021): 1-29..
